Graphics Gallery
Gale Rhodes
Chemistry Department
University of Southern Maine
Revised 2006/08/02
Learn how to use Swiss-PdbViewer. Work through sections 1-4 of
the Swiss-PdbViewer
Tutorial.

Topic: Glucose Catabolism
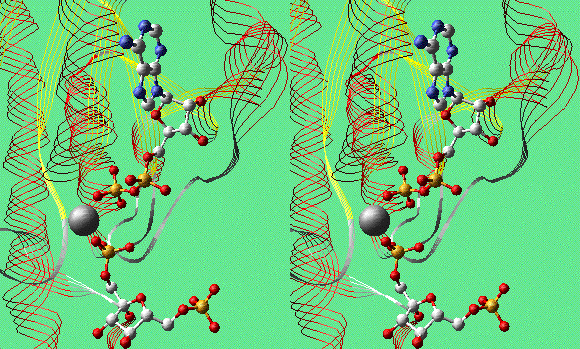
FBP, Mg2+, and ADP in PFK-1 (Convergent Stereo)

Examples:
Some Simple Carbohydrates
To review basics of glucose and glycoside structures, click
HERE for a Deep View project file
containing models of alpha- and beta-D-glucose, maltose, and
cellobiose.
Phosphofructokinase, Complex With Products and Allosteric
Effector ADP
Phosphofructokinase (PFK-1) is the pacemaker enzyme of glycolysis.
Here is a model of two subunits of the tetrameric enzyme from E.
coli: 1PFK.pdb.
At the active sites are the products, fructose-1,6-bisphosphate
(FBP) and Mg2+/ADP. The crystallographers attempted to
crystallize the enzyme with fructose-1-phosphate and ADP, which
should produce an inactive complex. To their surprise, the
electron-density map clearly showed FBP instead. They suggest that it
formed by reaction with contaminating ATP.
Think About It
- Two molecules of ADP are bound to each subunit. One ADP is
bound adjacent to FBP at the active site. The second ADP is far
away from this site. What is the role of the second ADP- binding
site?
- Examine the binding of Mg2+ to all four ADP's in
the dimer. Does the ion play the same role in all cases?
- The PDB file 2PFK.pdb is a model of apo-PFK. Get
2PFK.pdb and and superimpose one subunit of it onto 1PFK. Than
explore the conformational differences between the two
models.
- Search the PDB for other PFK models that will illuminate the
effects of substrate and effector binding (use PDB SearchLite and
search for "phosphofructokinase").
Glyceraldehyde-3-Phosphate Dehydrogenase, NAD Binding Site
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) catalyzes the
reversible oxidation of G3P by NAD+. Here is a model of
the dimeric GAPDH from E. coli: 1GAD.pdb.
Explore the NAD binding site. Find the catalytic side chain of
CYS149 near the nicotinamide ring.
Think About It
- Display the backbone of the protein and color by secondary
structure. What type of structure forms the interface between the
subunits?
- The nicotinamide ring of NAD+ can accept hydride
ion on either of its two faces. Enzymes that catalyze this
reaction are usually stereospecific for one face or the other. If
you view the nicotinamide ring with C-4 at the top and the amide substituent
to your right, the top face of the ring is called the A face, and the bottom
is the B face. Use SPdbV to determine whether GAPDH would direct
addition of hydride ion to the A or the B face.
- The N313T site-directed mutant form of GAPDH exhibits weaker
binding of NAD and lower catalytic efficiency. Look at the
position of ASN313 with respect to NAD. What kind of interaction
occurs between them? What role might ASN313 play in binding and
catalysis? If you use SwissPdbViewer, you can compare the mutant
with this model. Click here
to obtain the coordinate file for the N313T mutant from the
Protein Data Bank.
Pyruvate Decarboxylase (Yeast), TPP Binding Site
Pyruvate decarboxylase contains the cofactor thiamine
pyrophosphate (TPP). The active form of TPP is the conjugate base,
resulting from loss of a proton from C-2, the carbon between N and S
in the thiazole ring. Here is a model of the dimeric enzyme from
yeast: 1PVD.pdb.
Like many di- and tri-phosphates, TPP is bound to its enzyme as a
complex with Mg2+. Restrict your view to atoms within 7 or
8 angstroms of TPP in chain A to see the binding site for TPP.
Display the alpha carbons of chains A and B in differenct colors and
notice that the TPP binding site includes residues from both
subunits. The active sites in this dimer lie at the subunit
interfaces.
Think About It
- Can you find a nearby group that might be responsible for
deprotonating C-2 of TPP? What interactions position this group
for its role?
- What might be the role of the pyrophosphate/Mg2+
moiety of TPP?
- Would you expect a plot of rate versus substrate concentration
for this enzyme to exhibit square hyperbola or sigmoid shape?

Topics List
Biochemistry
Resources


![]()