Graphics Gallery
Gale Rhodes
Chemistry Department
University of Southern Maine
Revised 2006/11/07
Learn how to use Swiss-PdbViewer. Work through sections 1-4 of
the Swiss-PdbViewer
Tutorial.

Topic: Lipids and Membranes
Thanks for Eric Martz for preparing the
Mebrane Lipids and Ionophores files in PDB format.
They are part of a RasMol animation of lipids and bilayers, available
at the RasMol Home
Page.
For a full description of the bilayer models provided and
discussed on this page, see "Molecular dynamics simulation of a
bilayer of 200 lipids in the gel and in the liquid-crystal phases",
H. Heller, M. Schaeffer, and K. Schulten, J. Phys. Chem, 97,
8343-60 (1993).

Examples:
Membrane Lipids
Cholesterol, Phosphatidyl
Choline, and Lipid Bilayers (Deep View project file)
NOTES, Layer 1, cholesterol
- Cholesterol is amphipathic, having both polar and
nonpolar regions. What functional group constitutes the polar
region.
- Cholesterol is a normal component of cell membranes. How would
it be oriented in a lipid bilayer?
- Cholelsterol stiffens the outer parts of the phospholipid
tails, thus making the membrane less fluid.
- Counterintuitively, cholesterol also acts as an impurity,
lowering the "freezing" temperature of the membrane -- the
temperature at which the membrane undergoes a transition from the
fluid phase to the gel phase. So it lowers membrane fluidity, but
widens the temperature range over which the membrane remains
fluid.
NOTES, Layer 2, phosphatidyl choline (PC)
- Note the amphipathic nature of PC.
- Identify the fatty acids in these PC molecules. Each molecule
has two different fatty acids esterified to it.
- Display cholesterol and PC together. Approximately how would
their polar heads align if they were neighbors in a membrane?
NOTES, Layer 3, 40 PC molecules in a lipid bilayer, with water
molecules on one side.
- Compute hydrogen bonds and look for examples of H-bonds
between water molecules and PC head groups. Does the absence of
H-bonds involving the positively charged triethyammonium groups
surprise you? Explain.
- Note that all fatty-acid tails are shown with the same
conformation. This is an idealized model. A larger model like it
was used as a starting point for molecular dynamics simulations of
motion in lipid bilayers. To see the resulting nonidealized model
after 100 picoseconds of computer-simulated motion, click
here (NOTE: large file!).
This will add a fourth layer, called Fluid, to the display. This
model is probably much more like a real bilayer.
- Compare the idealized and "real" models as to 1) the variety
of hydrocarbon conformations, 2) thickness of the bilayers. Why is
the dynamics model thinner?
- For a smaller non-idealized bilayer model, see the gramicidin
model next.
Phospholipases
A
Competitive Inhibitor of Phospholipase A2
Click the link above to download a Deep View file derived from PDB
file 1POE.pdb
(click this filename to learn more about this protein directly from
its PDB entry).
Phospholipase A2 catalyzes hydrolysis of fatty-acid links in
phospholipids like phosphatidylethanolamine (PE), releasing fatty
acids from the 2-position of glycerol. Phospholipase model 1POE
includes a nonprotein ligand (designated GEL) which is an unreactive
analog of PE. This ligand binds tightly to the active site of the
enzyme, excluding its normal substrate, and thus reducing the rate of
ester hydrolysis. GEL is therefor called a competitive
inhibitor of phospholipase A2 -- it inhibits the enzyme by
competing with the substrate, the substance on which the enzyme
normally acts.
When you open this file, you will find GEL displayed with a van
der Waals surface, its neighbors (to 4 angstroms) displayed as stick
models, and the rest of the protein in ribbon.
NOTES
- Study the structure of GEL (dotted surface), and determine in
what ways it is like, and unlike, PE.
- Display hydrogen bonds between protein and ligand to see how
the polar groups of the ligand are stabilized by the protein.
- A PREVIEW: HOW ENZYMES WORK: Write the mechanism for
base-promoted hydrolysis of an ester. The product of the
rate-determining step is a tetrahedral intermediate resulting from
addition of hydroxide ion to the the ester carbonyl. Because this
intermediate is near the transition state on the
progress-of-reaction curve for this process, the transition state
must therefore be structurally very similar to this tetrahedral
intermediate.
- Compare the phosphodiester group at C2 of the glycerol to the
tetrahedral intermediate in ester hydrolysis -- they are very
similar in geometry and electronic structure. Thus, to this
enzyme, GEL looks much more like the transition
state in ester hydrolysis than like its reactant
(ester) or product (carboxylic acid). If an enzyme binds
strongly to, and thus stabilizes, the transition state of the
reaction it catalyzes, then it will lower the energy required to
reach the transition state and make the reaction faster. It
appears that stabilization of the transition state is at least one
aspect of how phospholipases catalyze ester hydrolysis.
- Inhibitors like GEL are called transition-state
analogues. Scientists can often use knowledge of enzyme
reaction mechanisms to design such inhibitors with the aim of
selectively blocking enzyme action. Some important drugs are
transition-state analogues for vital enzymes in pathogens.
- Protein-digesting enzymes know as serine
proteases hydrolyze peptide bonds by a mechanism similar to
that of ester hydrolysis by phospholipases. Look up the mechanism
of action of serine proteases in your biochemistry textbook. You
will probably find mention of an oxyanion hole at
the enzyme's active site. Can you find something similar to an
oxyanion hole in phospholipase A2? Look at how the negative
charges on the glycerol C-2 phosphate are stabilized.
To see this enzyme in the presence and absence of GEL, click
HERE.
- In layer 1POE, select GEL and then select and display its
neighbors to 4.5 angstroms. Select: Extend to other layers
to make the same selection in layer 1POD, which lacks GEL. Move to
layer 1 POD and press return to display only the selected
residues.
- Move back and forth between 1 POD and 1 POE (control-tab) to
see conformational differences between the two models. These
differences accomodate the substrate and move transition-state
stabilizing groups into position.
Ionophores
Gramicidin in Lipid
Bilayer
NOTES
- Gramicidin is an ionophore, or ion carrier. It make
membranes permeable to water and protons, but its permeability is
blocked by Ca2+ ions.
- Notice that the lipid side chains from the two monolayers
intermingle with each other. This model was produced by computer
simulation of the motion of all molecules, starting from an
idealized model like the one described above. This model is
probably much like a real bilayer.
- Notice the water molecules in the gramicidin channel.
- Rotate the model to look at it end-on. Only half the bilayer
needed to enclose the gramicidin molecule is shown in the
model.
- Notice that the ionophore is polar inside and hydrophobic
outside -- just the opposite of water-soluble proteins. What polar
groups make up the interior? How does the distribution of polar
and nonpolar groups compare with that of a water-soluble
protein?
Valinomycin
NOTES (read all before clicking the link at the end):
- Not a channel former like gramicidin, valinomycin wraps
potassium ions in a 36-member chain of amino- and hydroxy-acid
residues, giving it a hydrophobic covering in which it can
traverse the nonpolar regions of a lipid bilayer.
- To view the molecule with Deep View, click here.
Integral Membrane Proteins
Bacteriorhodopsin (PDB 1C3W)
NOTES
- Bacteriorhodopsin is a light-driven proton pump from a
halophilic bacterium. Its action produces
a pH gradient across the cell membrane. This gradient is the primary energy
source for all of the bacterium's cellular metabolism, including the production
of ATP for driving chemical processes like biosynthesis. The mechanism of
proton pumping is an active area of research. Bacteriorhodopsin is the best
understood transmembrane pump, but there are still many questions about
exactly how it works.
- "The
function of bacteriorhodopsin has been studied extensively by a
variety of structural, genetic, and spectroscopic methods and by
molecular dynamics calculations. The primary event, absorption of
a photon, causes isomerization of the retinal from the all-trans
to the 13-cis configuration. A series of intermediate events
follows, including deprotonation of the Schiff base and proton
transfer to Asp85. Another proton is subsequently released, the
Schiff base is reprotonated from Asp96, and a proton is taken up
from the cytoplasmic side. Thermal reisomerization of the retinal
to the ground state completes the photocycle." (from X-ray
Structure of Bacteriorhodopsin at 2.5 Angstroms from Microcrystals
Grown in Lipidic Cubic Phases, Eva Pebay-Peyroula, Gabriele
Rummel, Jurg P. Rosenbusch, Ehud M. Landau Science, Volume
277, Number 5332, Issue of 12 Sep 1997, pp. 1676-1681.)
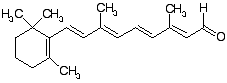
All-trans retinal
- Download a bacteriorhodopsin model by clicking on the PDB file
name above. Display the backbone as ribbon and color by secondary
structure. What is the main type of secondary structure in this
molecule? Why is this type of structure often found in
transmembrane regions of proteins?
- Display backbone and color by secondary structure succession.
Trace the chain from N- to C-terminus. Notice how this color
scheme makes it easy to follow the chain through the structure,
even if there are breaks in the sequence. Locate one such break
and use labeling to identify the residue numbers on each side of
the break. Why do you think some residues are missing from the
model? (Hint: the structure was determined by x-ray
crystallography.)
- Color backbone dark gray (so it id very faint against the
blace background), and color sidechains by type. By looking at the
distribution of sidechain types (charged, polar, nonpolar) on the
molecular surface, can you tell what parts of this molecule are
buried in the cell membrane and what parts are exposed at the
membrane faces?
- Notice a blue sidechain in the interior. What amino acid is
it? Why is it unusual to find this amino acid buried within a
protein? This sidechain is not its usual ionic self, however. It
is covalently attached to retinal, the free form of which is an
aldehyde. What type of chemical attachment is this, between an
amine and an aldehyde?
- Examine the contacts between retinal and the protein interior
(Use Select: Neighbors... to limit the display). What types
of interactions do you find? The fundamental event in proton
pumping is the absorption of a photon of light by retinal,
resulting in isomerization of one of its trans-double bonds to the
cis-isomer. The change in shape induces conformational changes in
the protein, which are thought to be essential to translocation of
a proton from one end of the molecule to the other, and hence
across the membrane. Would you describe the retinal pocket as
loose- or tight-fitting? Which would be conducive to coupling
isomerization of the retinal to conformational change in the
protein?
Bacteriorhodopsin "In Action"
Click HERE to download a DeepView
project file that depicts light-driven proton pumping by bacteriorhodopsin.
Before loading this file, make sure that DeepView loading preferences (Prefs:Loading
Protein) are set to load and show water molecules.
Crystallographers have determined the structures of many site-directed mutants
of this protein, including some whose spectral properties suggest that the
mutations have trapped them in conformations that represent intermediates in
the pumping process. I have assembled a set of these models to depict proton
pumping. The models appear in the project file in the order thought to represent
the stages of pumping. The protons are depicted by water models with dotted
surfaces. If you blink through this series, following a dotted ball of a specific
color, you will see it associate with the side-chain groups that have been
proposed as the proton-carrier sites. I emphasize that this is not an animation
of proton pumping. It is simple a series of crystallographic models upon which
one can illustrate the action of proposed proton carrying groups.
When the retinal model goes from purple to dark blue, it is depicting the
trans-to-cis isomerization that occurs on light absorption. The models shown
between that and the next light-absorption event represent pumping stages,
including at one point the re-isomerization of retinal to the trans- form,
which probably occurs spontaneously. At the cis-to-trans reisomerization, the
retinal model goes from yellow to light gray.
The first layer (named chain) of this file is a full model,
while all other layers are partial. For the most dramatic depiction, use the Wind:Layers
Infos command
to control the layers. In the cyc column, click the first
layer's entry repeatedly to turn it to a "+", which keeps in it view while
your blink through all other layers (they should have checkmarks in the cyc column).
In the chain layer, turn off display of all residues, leaving
only the ribbon. Use Prefs:Ribbons to display the ribbon as
a single strand. Now blink (hold down ctrl and press tab repeatedly)
to see the action. For the fastest pumping, hold down ctrl and
then just hold down tab.
OmpF Porin from E.
coli (Deep View project file)
NOTES
- Porins are channel forming proteins in the outer membranes of
gram-negative bacteria, mitochondria, and choloroplasts. They
permit entry of solutes smaller than 600kD, such as nutrients.
What is the signficance of the presence of porins in these
particular membranes, but not in the cytoplasmic membranes of
eucaryotes?
- The initial display in Deep View is ribbon backbone. Beta
strands are white, alpha helices are red, and loops are a
different color for each monomer of this oligomeric molecule. Also
on display are some detergent molecules that were part of the
crystallization medium. They are the same color as the chain with
which they are associated. What is the subunit composition of this
porin?
- What is its primary type of secondary structure? Color the
ribbon by secondary structure succession and trace the chain from
N-terminus (near the strand of darkest blue) to C-terminus
(reddest strand).
- Remove ribbons, and display the backbone of chain A. Color
backbone (only) CPK. Compute H-bonds and examine hydrogen bonding
in the outer shell of the structure. What are beta barrels and
alpha helices the two most common kinds of secondary structure
found in transmembrane regions of proteins?
- Color backbone dark gray (so it id very faint against the
blace background), and color sidechains by type (they are already
colored this way if you have followed these instructions
carefully). Study the types of residues that cover the porin
surface. By looking at side-chain types (charged, polar, nonpolar)
can you tell what parts of the outer surface are buried in the
membrane and what parts are exposed at the membrane faces?
- The transmembrane channel is constricted by what type of
structure? Look straight down into the pore. Use a 10-angstrom
slab to travel through the pore, as follows: Turn on slab, then
hold down <command> while you translate the molecule. In
effect this action slides the molecule along the z-axis through
the display slab, enabling you to see a moving thin section of the
structure. What are the predominant types of residues that line
the constricted part of the channel?
- Display all chains and look down the transmembrane direction
into the subunit interface. Then select and display only those
residues that are within 5 angstroms of the subunit interface
(Select: Groups Close to Another Chain...). This limits the
display to the residues that make interchain contacts. Now use the
slab to move through this interface. What types of interactions
hold the oligomers together?
- OmpF porin is weakly cation selective in permitting solutes to
cross the membrane. A similar channal molecule, PhoE porin, is
weakly anion selective. Obtain the original PDB for both porins
(2OMF
and 1PHO)
and compare them. Your biochemistry text might have additional information
about porins. If not, Google "porin".

Topics List
Biochemistry
Resources

