Molecular Graphics ManifestoAdd a substantial new skill to an overcrowded course? |
The first biochemistry course is already jammed with material, a daunting profusion of new terms, concepts, and skills that offer the potential of bringing the student to the level of primary literature after one year of study. Is this new tool of molecular graphics really important enough to take its place in the standard curriculum? Why not just teach the basic concepts with traditional materials, and relegate tools like graphics to the lab or to advanced courses?
The best reason is that hands-on graphics is a powerful enhancement to learning, particularly individualized learning. Using graphics as a tool reinforces textbook material and deepens understanding of it. There is powerful synergy in learning about proteins and learning simultaneously about how to represent and manipulate them with computer graphics. You can't learn about proteins without seeing the deep inadequacy of those lovely but often mostly cosmetic printed graphics in texts, as well as the limitations of computer-based ancillary materials, with their passivity and restricted interactivity. On the other hand, you can't learn to use graphics without seeing proteins and other complex biomolecules in a new and vivid way, and without discovering personal solutions to the problem of "seeing" new structural concepts. Learning is a highly personal process. Molecular graphics frees and invites individual students 1) to develop their own approach to clarifying and verifying what they read in the text, 2) to play actively with structure rather than merely to be shown it passively, and 3) to raise questions they can answer for themselveswith the tools at hand.
There are additional reasons that hands-on graphics should be added to the first biochemistry course. First, it strengthens other basic computer skills. Second, it enhances database and search skills, primarily by providing access to one of the most important bioinformatics databases, the Protein Data Bank. The PDB, in turn, is a natural portal to many other databases of structure, sequence, compound identification, and scientific literature. Finally, it is a powerful and marketable research skill. The genomics era is giving way to the proteonomics era, during which more detailed understanding of protein structure/function relationships must arise. We need to produce a corps of students who have a deeper understanding of proteins, and who are equipped with the tools to examine these marvelous machines, figure out how they work, and then to learn how to modify them for medical and industrial purposes.
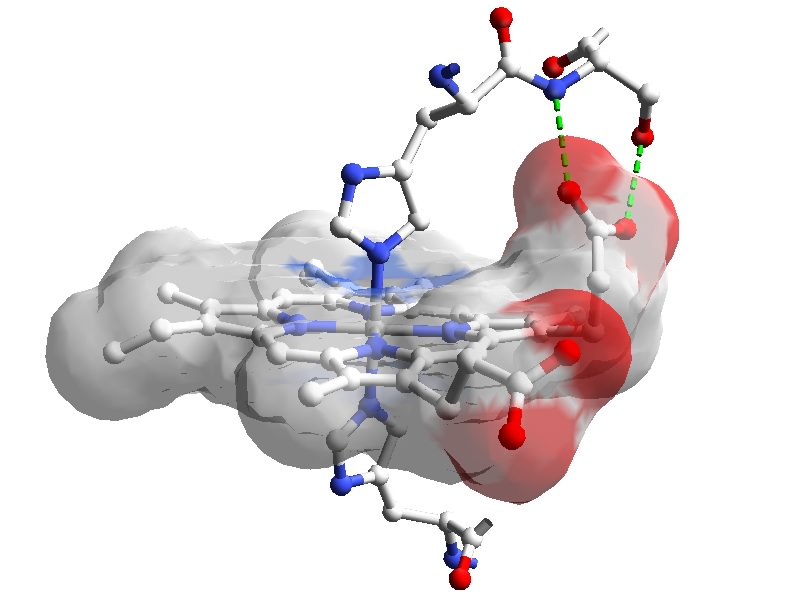
Figure: Heme with transparent molecular surface, and
neighboring residues in cytochrome b5. PDB 1CYO.
Created with Deep
View. Click image to enlarge.