Molecular Graphics ManifestoHow to Do It |
2. How can you add graphics without pushing out other topics?
As mentioned earlier, biochemistry courses are bulging at the seams with basic and essential topics, as well and new and exciting ones. In such courses, it is standard practice to take a sort of Sn2 approach to new course material; that is, if you add something, you must simultaneously displace something, especially if it is subject matter you cover in class. An efficient way to teach the use of a molecular graphics program is by use of a self-guiding tutorial in which students do most of the learning outside of class. In this year's course, after a brief introduction to Deep View in the classroom, students worked through a tutorial that guides them through the basic program functions, working toward more and more complex functions. Because students are new to the subject (proteins) as well as to the medium (graphics program), my tutorial teaches both graphics and structure. As students explore program functions, the tutorial guides them in seeing aspects of protein structure that they are concurrently learning from the text and class. As they explore protein structure, the tutorial guides them in learning how to manipulate and analyze structure gracefully, and in discovering the power that computer graphics puts into their hands.
I have found that an effective plan for a tutorial is to focus it on one small, well-understood protein, and give tutorial users an extensive guided tour of the protein using as many basic graphics functions as possible. The protein should contain all the basic structural elements: alpha helix, beta sheets, turns, and coil. The specific model of the protein under study should also contain a cofactor, inhibitor, or other bound ligand, allowing exploration of protein-ligand interactions. Even better, the protein should be covered in some detail in the course text, or studied in the lab, so that its role and mechanism of action will eventually become familiar to students. Students carrying out the tutorial thus become familiar with this protein and with how it illustrates various aspects of protein structure, and at the same time they learn how to manipulate it and to focus on its nature -- for example, its overall shape and dimensions, its secondary structural elements, the distribution of its charged, polar, and nonpolar groups, and the details of its interaction with ligand.
The subject of my tutorial is an old standard: PDB file 1HEW, a model of chicken egg-white lysozyme in complex with the competitive inhibitor tri-N-acetylglucosamine. This enzyme is usually among the first discussed in the introductory enzymes chapters of biochemistry texts, so information about its structure and action is readily available to students.
My Deep View tutorial is not specifically designed for my course and for my students, but for anyone who wishes to learn Deep View as a tool for learning, teaching, or research. The tutorial is on the World Wide Web (see Resources), and has been widely used and thoroughly tested by students, teachers, and researchers. Because the tutorial is for general use, my students may have to make some adjustments for their own computers or internet connections, which is good experience for dealing with learning tools on the World Wide Web. Carrying out the tutorial is a step toward independent use of the Web for learning.
If you teach graphics by way of a self-guiding tutorial, you can minimize the classroom time devoted to the subject, and thus displace a minimum of other classroom activities..
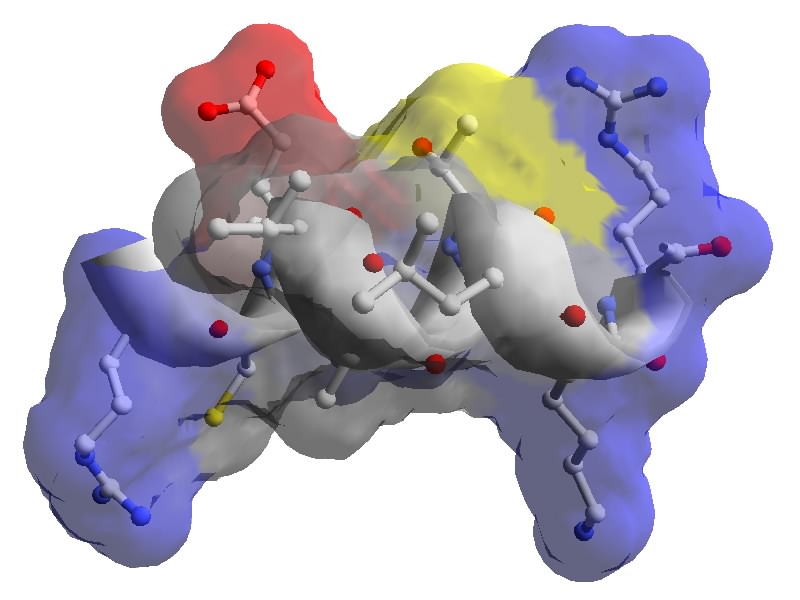
Figure: Amphpathic helix in human lysozyme. PDB 1LZS.
Created with Deep
View. Click figure to enlarge.