In this section, you will learn to study models that contain more than one protein chain. For this section, obtain the file 3HHB.pdb from the PDB.
Start DeepView and open 3HHB.pdb, a model of deoxyhemoglobin.
Color: Chain
Many proteins are oligomeric -- composed of more than protein
chain, or subunit. DeepView colors each subunit a different color.
This command is a quick way to find out how many chains are in a
newly opened PDB file.
Click on the Control Panel to activate it. Notice that letters A in the first column, the chain column. Click on any A, then press return. Clicking in the chain column selects the entire A chain. Scroll down to the end of the list of A-chain residues and click on any B, then press return. Chain B appears and chain A disappears. The chain column provides for quick and easy selection and deselection of entire chains. If the protein has only one chain, this column is empty. Clicking in an empty chain column selects the whole model.
Chains A and B in this model are the alpha and beta subunits of hemoglobin. You may be aware that hemoglobin consists of four subunits, two identical alpha and two identical beta. Some PDB files for oligomeric proteins contain only the unique subunits -- in this case, one each of alpha and beta. Later, you will build the additional subunits. But first, you will examine the interface between the subunits.
Color the chain A cyan (between blue and green on the color wheel) and the B chain magenta (between red and blue).
Select: Groups Close to another Chain...
Use this dialog to select groups that are within 5
angstroms of another chain, and click OK. This amounts to
selecting the residues within 5 angstroms of the subunit interface.
Press return to eliminate other residues.
Study the interactions in the subunit interface as follows. Color the selected sidechains (not backbones) by type, and then compute H-bonds. Zoom in for a close examination. The backbone colors allow you to distinguish chains A and B, while sidechain colors suggest the types of interactions. Red side chains (negative) near blue (positive) suggests ionic interactions. Gray near gray implies hydrophobic interactions. Green dotted lines indicate H-bonds. Do you see any interchain H-bonds? Measure any potential ionic or hydrophobic interactions. Slabbing and showing surfaces of selected groups may help you to see more about how the subunits interact.
When you are finished, redisplay the full model. Reset the Control Panel color menu to color backbone + sidechain, and color the model CPK. Turn off the display of H-bonds.
If you want to examine A-A or B-B interactions, you need a model of the entire tetramer. If the proper information is provided in the PDB file, DeepView can help you build the full model. In the remainder of this section, your will build the full tetramer from two copies of the file 3HHB.pdb.
File: 3HHB.pdb (look for this file name
near the bottom of the File menu)
This command takes advantage of DeepView's list of recently used
files. You have just used it to open a second copy of 3HHB. The two
models are superimposed, so it appears that nothing new has been
added.
Wind: Layer Infos
The Layer Information window allows you to control the display
and properties of multiple layers.
Color: Layer
<ctrl> - <tab>; <ctrl> - <tab>,
<ctrl> - <tab>
You are cycling or blinking between the two models, which are now
colored yellow and blue.
These two models are called layers. On the Layer Infos window, both layers are listed as 3HHB. Blink to make the second 3HHB visible (check mark in vis column) and active (layer name red). Now you will rename the second 3HHB so you can more easily tell the two layers apart.
Edit: Rename Current Layer...
Name the new layer 3HHBA'B' and click OK.
Blink between the two layers. Stop with the new layer active -- red in the Layer Infos window. The name 3HHBA'B' should also be displayed at the top of the Control Panel.
Make the Control Panel active and click on any A in the left-most column. You have selected all groups in chain A.
Edit: Rename Current Layer...
Tab into the box labeled Rename Chain of Selected Groups,
type C, and click OK.
In the 3HHBA'B' layer, select chain B.
Edit: Rename Current Layer...
Tab into the box labeled Rename Chain of Selected Groups,
type D, and click OK. You have renamed the A and B chains of
the new 3HHBA'B' layer so that when you merge both files into one,
the four chains will be named A and B (the first alpha/beta dimer), C
and D (the second dimer). Now you will rotate the CD pair into the
orientation of the second alpha/beta dimer.
Select:All
With all groups in the new layer (3HHBA'B') selected, click the
file icon on the graphics window (beside the little earth icon) to
display the PDB file. Scroll down to the beginning of the ATOM lines.
The last three lines, labeled MTRIX, before the first ATOM line give
the information that DeepView needs to transform the coordinates of
your CD dimer into the coordinates of the second, or A'B' pair. All
you need to do is click anywhere in these three lines, and click
OK on the dialog box. Then close the file window, and you will
find the current layer in its new orientation, with the original
layer in the old orientation. Together, they make a full
tetramer.
Blink between the layers to see their relative orientation. In the Layer Infos window, put checkmarks in the vis column beside both layers to see them together. Next you will combine them into a single layer, and then save the tetramer in a new file.
<shift> - Select: All (selects all in both layers -- recall that shift applies Select commands to all layers)
Check to be sure that all residues in both layers are selected.
Edit: Create Merged Layer From Selection
This operation will take a few seconds. Notice that you now have a third layer, named "_merge_" . Blink to make _merge_ the active the layer.
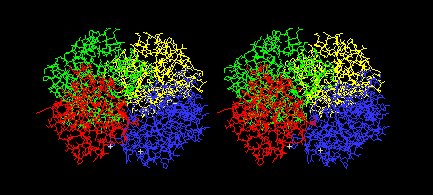
Color: Chain
You should see the four chains of the tetramer in different
colors, like this:

File: Save: Layer
Save the _merge_ layer in a file named 3HHBTet.pdb.
Close all files and then open your newly saved file. Rotate and study the way the four oligomers fit together.
Select: Groups Close to Another Chain
Use the dialog that appears to select groups that are within 5
angstroms of another chain, and click OK.
<return>
Now you can study all of the subunit interfaces of the tetramer,
using the same tools you used earlier to study the AB
interface.
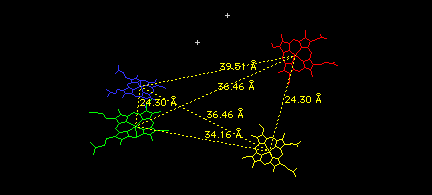
Finally, look at the relative positions of the four hemes (and two phosphate ions) in the tetramer as follows:
Select: Group Kind: HETATM
<return>
You should see only the four hemes and two phosphate ions. Use the measurement icon on the graphics window to measure all the distances between iron atoms in the various hemes. When you have measured them all, the six yellow lines will form an elongated tetrahedron, with its shortest sides connecting the two alpha/beta dimers you have combined to build this tetramer.
Close all files.
![]()
![]()
To Next Section: 11. Comparing Proteins