TEACHERS: This assignment provides a means to assess your students' command of DeepView and the basics of protein structure. It is appropriate for first-semester biochemistry students who have completed an introduction to protein structure and Sections 1-6 of the DeepView Tutorial. Click here for a list of appropriate PDB files for this assignment.
OTHERS: Use this assignment to review tutorial ideas, or to test your command of the program. When in doubt about how to complete a task, go back to the tutorial to see how you accomplished the same task there. If you have worked through the first six sections of the tutorial, you have done everything that this assignment asks.
Revised May 30, 2008.
This assignment gives you the opportunity to demonstrate your ability to use the basic functions of DeepView. Pick a protein to study from the list below. You will prepare some specified views of your protein, save them as DeepView coordinate files, and turn them in the files for evaluation. Finally, you will write a brief description of the protein, some of its structural features, and its interaction with its hetero group.
Click here to see a list of appropriate proteins. Each student in a class should work with a different file. All assigned proteins function as single chains and contain a hetero group: a cofactor, prosthetic group, or inhibitor.
If you prefer, you may choose and study a protein of particular interest to you, but it must function as a single chain and contain a hetero group. In addition, it must be a protein that no one else in the class has already chosen. Proteins with more than 250 residues can be very difficult for graphics beginners, so smaller is better. Please ask your instructor to approve your choice if it is not from the assignment table.
PLEASE NOTE: YOU CANNOT RECEIVE CREDIT FOR WORKING ON A PROTEIN ASSIGNED TO SOMEONE ELSE OR NOT APPROVED FOR YOU.
Whether you accept an assigned protein or find your own, obtain your file from the Protein Data Bank using your browser.
Using what you have learned from Sections 1 through 6 of the DeepView tutorial, create the views described in the following list. Save each view with a filename in this format: xxxx#.pdb, where xxxx denotes the PDB file code for your protein, and # is the view number expressed as two digits (example, for View 3 using 1hew, the file name would be 1hew03.pdb and view 10 would be 1hew10.pdb)
Once you have constructed the required view save it as follows:
File: Save: Layer...
The familar file-saver dialog box appears. Navigate to the
location where you want DeepView to store the file (if you are using
a USM computer, save them to the desktop and later transfer them to
your own disk using the file-copy functions of your operating system
-- see this warning),
type in the appropriate file name, and click Save. At the end
of this exercise, you will combine all of your files into one DeepView Project File (instructions below).
If your protein contains two or more identical chains (this will be noted in the proteins list), create a working file containing only the one chain that you plan to study, with its hetero group. The command File: Save: Selected Residues Only... is useful for this purpose. Then use this file as the starting point for preparing all the required views. This will make it much easier for you (and your reader) to restrict all views to one chain and its associated hetero group. If there are hetero groups in addition to the one listed in the table of assigned files, include in your working file only the hetero group listed in the table. Finally, do not leave water molecules on display in any of your completed views.
DeepView saves coordinate files in their original orientation if you use any of these commands:
DeepView saves coordinate files in their current orientation if you use this command:
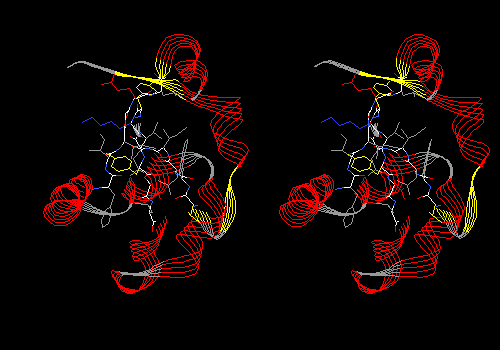
Following is the list of views to make and save. Each is accompanied by an example -- a convergent stereo snapshop of cytochrome b5 (PDB 3B5C) in the assigned view. Cytochrome b5 is a liver redox protein that contains a heme prosthetic group.
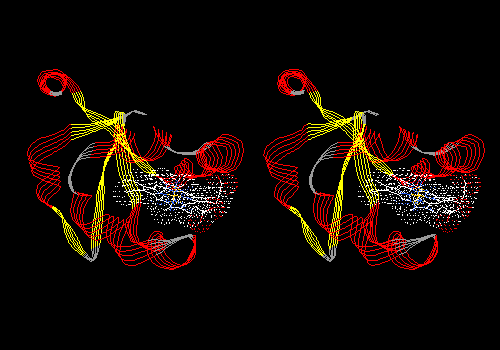
View 1 (filename xxxx01.pdb): Protein in ribbon model only, colored by secondary structure. Ligand colored CPK with dotted van der Waals surface.
Example:
View 2 (filename xxxx02.pdb): Detail of an alpha helix (if your protein contains no alpha helix, you can skip to View 4). main chain and side chains colored by type. Compute and display hydrogen bonds, and arrange the view to emphasize hydrogen bonding (Hbonds will not be saved). Label the terminal residues of the helix.
For your written description, note the distribution of polar and nonpolar residues around the helix.
Example:
Note that most of the residues pointing to the right and back are polar. Three of the residues pointing left are nonpolar (gray). NOTE: In all of your views that contain color by type, the backbone of each residue will be colored the same as the side chain, so your views will not look exactly like the ones shown as illustrations.
View 3 (filename xxxx03.pdb): Same helix as in View 2 in the context of entire protein. Remainder of protein in ribbon model from View 1. No ligand. No labels. Be sure that ribbon is NOT shown for residues in the helix under examination (Select: Inverse Selection may be a handy command in making the ribbon part of this view).
For your written description, note the location (surface, buried) of the helix. In the description, relate the location to the distribution of residue types as shown in View 2.
Example:
Note that the helix is on the surface, with much of its surface exposed to solvent. Thus most of its sidechains are polar. The three nonpolar side chains are on the interior of the helix. In later views, you can see that these side chains are in contact with the heme prosthetic group.
View 4 (filename xxxx04.pdb): Detail of 3 or 4 strands of pleated sheet (if your protein contains no pleated sheet, skip to View 6). main chain and side chains colored by type. Labels on termini of each strand.
For your written description, note the distribution of polar and nonpolar residues on opposite sides of sheet.
Example:
Note that both sides of this three-strand sheet contain mostly nonpolar side chains (gray -- tyr is yellow, but is also mostly nonpolar). Some residues at the top and bottom edges are polar. The next view shows why.
View 5 (filename xxxx05.pdb): Same strands as above in contex of entire protein. Remainder of protein in ribbon model as in View 1. No ligand. No labels. No hydrogen bonds. Be sure that ribbon is NOT shown for residues in the sheet under examination.
For your written description, note the location (surface, buried) of the sheet. Relate the location to the distribution of residue types on opposite sides of the sheet.
Example:
Note that the side of the sheet facing away from the viewer is buried and contains mostly nonpolar (gray) sidechains. Surprisingly, several side chains on the nearer, exposed face of the sheet are also nonpolar. This form of cytochrome b5 is actually a fragment cut from a larger protein. In the intact protein, most of this sheet if buried, and thus most side chains on both sides of the sheet are nonpolar. The sheet forms the back of a pocket that holds the heme.
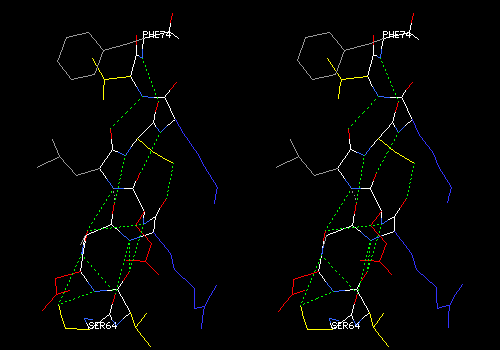
View 6 (filename xxxx06.pdb): Ligand with residues that lie within 5.0 angstroms of the ligand. Ligand colored CPK, main chain and side chains colored by type. Ligand labeled.
Compute H-bonds and explore hydrogen bonding between ligand and protein. For your written description, make a list of hydrogen bonds that link the hetero group to the protein. For each bond, list donor and acceptor atoms, using PDB atom designations the DeepView uses in its labels. If any hydrogen bonds from or within the hetero group appear to be erroneous, mention them in your description. (You can remove them (Build: Remove H-Bond...) for purposes of making a publishable view, but they will reappear the next time you compute H-bonds.)
CRITERIA FOR IDENTIFYING HYDROGEN BONDS AND SALT LINKS: Remember that hydrogen bonds involve two electronegative atoms that share one hydrogen atom. In proteins, hydrogen bonds involve mostly N and O, but never C. A thiol sulfur of cysteine can form a weak hydrogen bond with N or O, but not with another S. Hydrogen atoms are not visible in electron density maps from protein crystallography, so they are not shown in protein models. Therefore, we must infer the presence of a hydrogen bond when we find a pair of appropriate atoms within about 2.8 angstroms of each other. In addition to chemical criteria (one atom must be of a type that is normally protonated, one atom must be N or O, and the other must be N, O, or S) and distance criteria (2.8 ± 0.5 angstroms), the atoms must satisfy geometric criteria for hydrogen bonding. The hydrogen is involved with a bonding orbital on one atom, and with a nonbonding orbital on the other, so a line extending from one hydrogen-bonded atom to another must make either a tetrahedral or trigonal angle with the other covalent bonds at both atoms. Salt bridges, or electrostatic interactions, must involve atoms of opposite charge. The atoms can be in almost any orientation to each other, at distances near the van der Waals contact distance of about 3 Å.
Example:
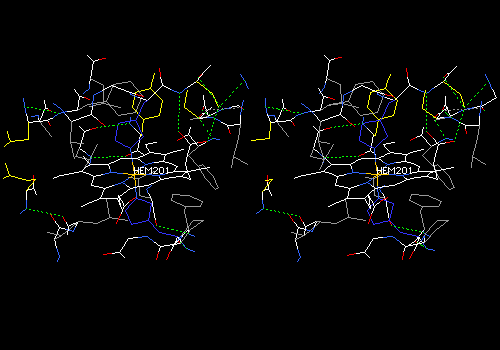
In the foreground, a heme carboxyl makes two hydrogen bonds with a serine reside (yellow side chain). One bond is to the main-chain NH, and the other to the side-chain OH of serine. DeepView draws some erroneous hydrogen bonds between the histidine ring about the heme iron to the heme nitrogens. These bonds have been removed. All this is hard to see without stereo.
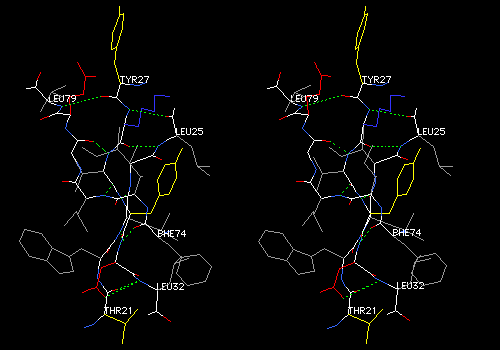
View 7 (filename xxxx07.pdb): Same as View 6, but van der Waals surface dots on all atoms. Ligand and its surface colored a bright shade of purple (brighter than the example below). main chain AND sidechains colored CPK. While View 6 emphasizes hydrogen bonding and polar interactions between protein and ligand, View 7 allows you to find nonpolar or hydrophobic interactions. These forces are discernable as dotted surfaces that make intimate contact with each other, like that between the which PHE58 and the purple heme group in the figure below.
Explore View 7 by slabbing (Display: Slab). Adjust slab depth to 5 angstroms in the Display Window Attributes dialog. Hold down <Command> while translating to move the slab forward and back. Find and save a view in which some specific ligand-protein contacts are evident. The ligand will probably be cut in cross section in such a view. Add some labels to show which residues are involved in contacts. Note that DeepView does not save slab settings, but slabbing will help you to find the interactions.
For your written description, list residues that make non-polar contacts with the ligand.
Example:

This cross-section of the nonpolar heme group (purple) shows surface contacts with several nonpolar sidechains.
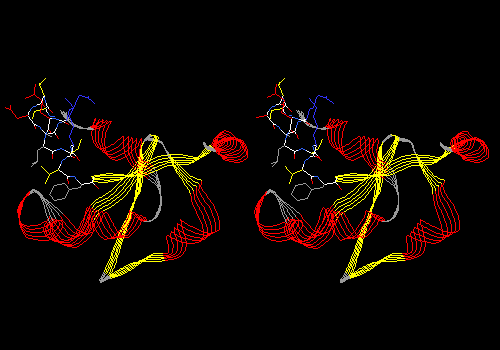
View 8 (filename xxxx08.pdb): Ribbon display of main chain, colored by secondary structure succession. Ligand in CPK with dotted van der Waals surface. Side chains (but no main chain) within 5.0 angstroms of ligand in CPK. This is similar to the first view of a protein you might see in a textbook.
NOTE: Due to a bug in some versions of DeepView, the setting Display: Show Sidechains even when Backbone is hidden is not saved. In written description, instruct the reader to turn on this function.
Example:
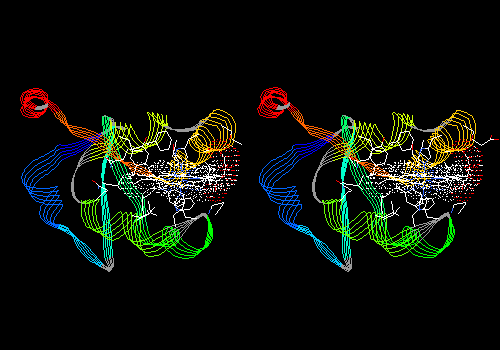
To see something lovely, try rendering this view in 3D (Display: Render in 3D; then Display: Render in Solid 3D). You can play with the appearance of rendered views by adjusting settings in Prefs: 3D Rendering and Prefs: 3D Lights. Press <Command>-3 again to put you back in normal viewing mode.
Rendered on a Macintosh computer, the model of cytochrome b5 looks like this (after the inevitable degradation of conversion to a GIF file):
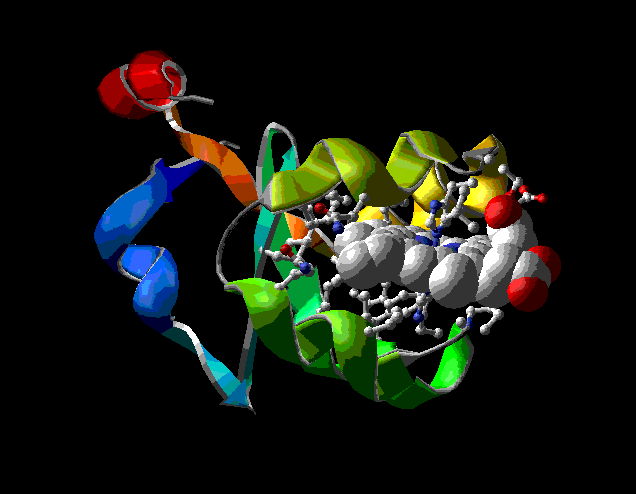
You can save pictures like this for use in reports and web pages with the command File: Save: Image.
NOTE about image-file formats: On a Mac, the result is a PICT file; on a PC, the result is a .bmp, .tiff, or (ugh!) .tga file, depending on settings of the operating system. The .tga file is hardest to use, and usually require conversion to some other format for use in Word. If you have trouble importing the file into Word, import it into a file-conversion program () or image-processing program (like Photoshop or Canvas) and File:Save As...: in JPEG or GIF format.
For more information on using these tools, see Making Images for Publication.
View 9 (filename xxxx09.pdb), and additional views, if desired: One or more additional views of your own design, aimed at illustrating unique features or specific functions of your protein. Be creative!
For excellent additional views, get to know your protein, and search it for some of the special structural features mentioned in your text. Search your protein for structural features in the following list, and if you find one or more, create views to show them off. For help with recognizing each feature, look it up in the index of your text. Most of these features are discussed in any biochemistry textbook, usually in the chapter that introduces three-dimensional structure of proteins.
At the end of your protein description, include separate captions for views #9 and higher, and for the solid image.
Create a cover sheet for your report by using Display: Render in Solid 3D to turn a revealing view of your protein into a striking image. Use Save: Image or Save: Image (Stereo) to turn this image into a file that you can import into Word to make a cover sheet. See earlier note about image file formats.
Before carrying out this step, look below find the Description and Structural Analysis project assigned for this year. Create any additional views need for your project. Then proceed with testing and combining your files, as follows:
Test-run each of your .pdb files by opening them in DeepView. Open them in the order created, and do not close each file before opening the next. You will have all nine or more files loaded at once (note: you can't reload image files).
<ctrl>-<tab>, <tab>,
<tab>, ... (hold down the ctrl key, and press tab repeatedly.
This operation is called "cycling" or "blinking". Each time you press tab (while
holding down ctrl), you turn off display of the currently displayed layer,
and turn on display of the next layer on the list. Thus you can move easily from
one of your views to the next. Blink through all your views and make sure that
each has the correct settings (review instructions, above, for each view). Correct
settings as needed. As you make these changes, you are not changing any of your
saved files. If you make any changes in a layer, File:Save:Layer... to
update that file.
If all views are OK, and you do not plan to combine them, they are ready to turn in with the other required materials. If you want to combine them into a single project file, read on.
Wind: Layer Infos
This command opens the Layer Information window, which
allows you to manage multiple files. Each file that you load becomes
a separate layer.
This window also allows you to move one layer without moving the others. When you rotate or translate, all layers normally move, whether they are visible or not. But if you remove the checkmarks from all but one layer in the "move" column, then only the checked layer will move. So if you wanted each layer in its own orientation, you could blink to it, turn off movement in all other layers, place it the way you wish, and then uncheck its "move" column. Then you could blink to another layer and repeat. It is probably easier to follow a tour of a protein if each layer is in the same orientation. But if you want each view to have its own orientation, use these tools to accomplish that. Remember that anyone who opens your project file and moves the model will be moving all your layers.
When all views are as you want them, it's time to save the entire project, and DeepView will save all files in their current orientation. Just follow these instructions:
Save: Project
Name the project file "#xxxProj.pdb", and save it to your
desktop. Quit DeepView, start it again, and open your project
file. Do a final check of all views to be sure they are as you want
them.
If you plan to hand in your project file on a disk (not the preferred method, but allowable: see Turning In Your Work), transfer it from desktop to disk using the file-copy functions of your operating system.
WARNING: Do not save files directly from DeepView to small disks, such as floppy disks. DeepView fails to inform you of save errors, which may occur if your disk does not have enough room. If, upon reopening the file from disk, you get the message: "File ignored (Either it is not a valid pdb file or it contains only a carbon alpha trace)" it means that the file was not saved properly or completely. Save project files to the desktop first, then check them, then transfer or email them.
![]()
Your instructor will ask you to complete this assignment with one of the following Description and Structural Analysis requirements:
To complete this assignment, search your protein for features given in the list Motifs and Less Common Structural Features in Proteins, above. Create at least two additional views (call them views 9, 10, and so forth) to illustrate such features in your protein.
Next, using Word, Appleworks, or Pages, write a caption for each View. Assume that your reader
In the caption for View 1, give the name and a brief description of the function of your protein. In the other captions, include instructions for changing the view, if necessary, to see features described in the caption, as in these examples, for a hypothetical enzyme called gigatuschin:
View 1: Gigatuschin is an enzymatic growth factor, over-expression of which causes rodents to have very large hips and thighs. The enzyme requires a cofactor, butylubiquitusch (BUT), to exert its effect. The protein is shown in ribbon diagram, colored by secondary structure. The cofactor is shown as a wireframe model with dotted van der Waals surface.
View 2: Alpha helix involving residues 56-66 in gigatuschin, in CPK colors. Side chains are colored by type. This helix lies on the surface of the protein, and as expected, it is amphipathic, with the outside face of the helix carrying predominantly polar side chains, while the inside face is mostly hydrophobic.
View 6: Detail of the BUT binding site. Compute hydrogen bonds, and note that BUT is held by hydrogen bonds involving serine-174 and asparagine-45 of the protein. In addition, the quinone ring of BUT fits into a hydrophobin pocket formed by aromatic residues tyrosine-57 and tryptophan 61, whose side chains stack above and below the ring. Arrange the view to see these rings edge on, and then use clipping to see the details of stacking.
Provide these captions as a simple list in a word-processor file. If your word processor is not Microsoft Word, Appleworks, or Pages, save the file in .rtf format, and make sure it looks OK when you reopen it.
To help you write informed captions about your protein and its hetero group, here are some potential sources of information:
End of DSA-1 project.
If your instructor assigned DSA-1, skip to Turning In Your Work.
To complete this assignment, write a brief (no more than one page, single spaced) description and structural analysis of your protein, illustrating it with the views saved in your project file, and presenting the information you were asked to gather as you made each view. Assume that your reader knows how to use DeepView, and imagine that your reader will open your project file and move from view to view as you discuss your protein. Assume that your reader's knowledge of protein structure is similar to yours. At the end of your description, don't forget to include captions for additional views, if any, of your protein.
This description should be an example of your best writing. Prepare it with the same care you would take in a writing course. Make your ideas clear and insightful; organize your paragraphs coherently, and put them in sensible order; make your sentences logical and grammatically correct; choose your words thoughtfully and spell them correctly. Cite sources of factual information about your protein, using the citation formats you see in PubMed. Your instructor will reject descriptions that do not meet these guidelines, or that are not organized as described below. Acting just like a the editor of a scientific journal, your isntructor will not edit a poorly written, carelessly proofread, or inappropriately organized description -- he or she will simply return it for rewriting.
Turn in your description as a word-processor file, in Word, Appleworks, Pages, or RTF format.
Give the description an informative title, and list yourself as the author.
In the first paragraph, give the name and source (organism) of the protein, and then describe its function and the function of its hetero group. If it is an enzyme, give the reaction that it promotes, drawing molecular structures of substrate(s) and product(s). (ISIS/Draw is an excellent, free program for drawing molecular structures in a word-processor document.)
In the second paragraph, describe the structure, including 1) the number of chains and number of residues in each, 2) major secondary structural elements and the residue numbers that they encompass, and 3) the location among your structural elements of the hetero-group binding site, all the while referring to the appropriate views.
In the third paragraph, describe the binding of the hetero group to the protein, referring to the appropriate views. Include description of the protein-hetero H-bonds, as well as hydrophobic and other types of interactions. Be sure to look carefully at all groups within 5 angstroms of the hetero group so that you do not miss important interactions.
Model your written description after the introductory description of any protein (for example myoglobin) in a biochemistry textbook. Write this description with a word processor, print it, and save it to the same location as your project file. Word processor programs allow you to save files in various formats. Please submit your file in one of the following formats: AppleWorks, ClarisWorks, Microsoft Word, RTF, ASCII text, or text.
NOTE: If aspects of your prepared views are not fully saved (like Hbonds, display of sidechains without main chain, or color schemes), instruct your reader to turn on the desired functions when you first refer to that view.
Here are some potential sources of information about your protein and its hetero group:
End of DSA-2 project.
![]()
When you have completed your project file and description file, submit them to your instructor, in one of these ways (in order of preference, best method first):
Please be sure that all files are in your instructor's hands by the deadline given in the syllabus. If you are uncertain about whether your files are readable, submit test files ahead of time. Instructors should check them and let you know immediately if they are usable. For full credit, your instructor must submit all files in usable condition before the deadline.
![]()
If you are interested in a particular protein that is not on this list, search the Protein Data Bank to see if its structure is known, and if there is a model containing a hetero group. If so, ask your instructor to see whether the model is reasonable and appropriate for this assignment.
|
|
|
|
| Student Name |
Phthalate dioxygenase reductase |
FMN |
|
|
Adenylate kinase |
ATP-analog inhibitor AP5A in two conformations |
||
| Student Name |
DNA (cytosine-C5-)-methyl transferase |
S-adenosylmethionine |
|
| Student Name |
Citrate synthase |
Coenzyme A |
|
|
Transketolase (2 chains -- use one) |
Thiamine pyrophosphate |
||
|
Aspartate aminotransferase |
Pyridoxal phosphate |
||
| Student Name |
Adipocyte lipid-binding protein |
Oleic acid |
|
|
Avidin (2 chains -- use only chain B and BTN401). |
Biotin |
||
| Student Name |
Phosphoenol pyruvate carboxykinase |
ATP |
|
| Student Name |
Methionine synthase (2 chains - use one) |
Vitamin B12 |
|
| Student Name |
H-protein of glycine cleavage system |
Lipoic acid |
|
Student Name |
Cellular retinol-binding protein |
Retinol |
|
| Student Name |
Dihydrofolate reductase (2 chains -- use one) |
Folic acid |
|
| Student Name |
Flavodoxin |
FMN |
|
| Student Name |
Acetyl-CoA carboxylase |
Biotin |
|
| Student Name |
Allophycocyanin (2 chains -- use one) |
Phycocyanobilin |
|
| Student Name |
HIV-1 protease (2 chains -- use both) |
Sulfamide inhibitor AHA006 |
|
| Student Name |
Alpha-thrombin |
Peptide-analog inhibitor |
|
|
Dihydrofolate reductase |
NADP |
||
Student Name |
Lactate dehydrogenase |
NAD |
|
|
Glutathione reductase (2 chains -- use one.) |
FAD |
||
| Student Name |
T-cell signal-transduction molecule SAP (two chains [A & C] and two polypeptide ligands [listed as chains B & D] -- work with only chains A (your protein) and C (the ligand). |
Phosphopeptide binding target |
|
| Student Name |
Ras protein catalytic domain |
GDP |
|
| Student Name |
ATP-binding cassette (ABC) transporter protein |
Mg-ADP |
|
|
Biliverdin IX Reductase |
NADP and biliverdin (analyze binding to either one) |
||
|
Rab4A, Ras-related protein |
GDP |
||
| Student Name |
Cytochrome C-500 |
Heme |
|
| Student Name |
Peptide deformylase |
Sb-543668, a potential pneumonia drug (2 superimposed models in alternative conformations) |
|
| Student Name | 1DB1 | Nuclear receptor for vitamin D | Vitamin D |
| Student Name | 1ASH | Hemoglobin from the nematode Ascaris suum | Heme |
| 1S8G | Phospholipase A2 | Dodecanoic acid | |
| Student Name | 2CEO | Thyroxine-binding globulin | Thyroxin (see note at top of table: files contains two models) |
| Student Name |
GFP-like fluorescent protein |
Fluorophore, made from modified residues 68, 69, & 70, listed in file after residue 67 as as CR768 (covalently bound, but peptide bonds might not be drawn -- find them) |